Understanding of host-pathogen interaction using extracellular vesicles and
their suitability in disease diagnosis and monitoring
Zoonotic diseases are of key relevance in comparative medicine since they impact both human and animal species. Early detection and accurate understanding of host-pathogen interactions are important in devising preventive and therapeutic approaches of zoonotic and any other pathogen mediated diseases. The early host-pathogen interactions in gastrointestinal pathogen-mediated diseases are very complicated. Often, there are no other ways to detect the early disease events than identifying the pathogen itself. Both pathogen and host are communicating with each other during the stages of disease establishment using different modes including chemokines, other bio active messengers and immune modulators. Interestingly, recent literature suggests the involvement of EVs in host-pathogen communications in many diseases. Thus, in the current project we are trying to extracts EVs in a non -invasive mode of sampling using feces / dung/ excreta as the EV source. We will enrich EVs from both cows and calves of healthy and Cryptosporidium spp. and Giardia spp infected animals and subject it to advanced genomics and proteomics analysis. Moreover, we will try to employ a novel metagenomic approach to understand the microbial dynamics (microbiota) as well. This analysis will let us understand specific biomarkers in EVs involved in the host-pathogen interactions and enable us to develop new diagnostic biomarkers of diseases and understand how some animals develop resistance to pathogens as well.
Project background
Since people are interacting with animals in their day-to-day life, most zoonotic diseases are commonly found among them. According to the Center for Disease Control and Prevention (CDC, USA), out of 10 known infectious diseases, 6 are transmitted from animals. These types of zoonotic infectious diseases could badly affect not only human and animal health but also the productivity of livestock. Cryptosporidium and Giardia represent the one of the most common intestinal parasites in Eastern Europe and are common in humans and animal species, especially in bovines. Their transmission is typically achieved through the faecal-oral route, by contact with contaminated stool. Cryptosporidium spp. and Giardia spp. have been respectively ranked as the sixth and 11th most important food-borne parasites globally. According to the review by Plutzer et al (2016), upgrading and making the diagnosis/detection procedures are not sufficient to obtain enough information regarding these infectious diseases in Eastern Europe since the variation of diagnostic methodologies, reporting practices throughout the region and not having enough investigation reports.
Conventional diagnostic techniques such as microscopic methods, staining methods, antigen detection tests, immunological/ immunofluorescence assays and nucleic acid based diagnostic methods such as polymerase chain reaction (PCR) are currently being used for the diagnosis of many bovine intestine related/associated infections including cryptosporidiosis (by Cryptosporidium sp.) and giardiasis (by Giardia sp.). Moreover, understanding the exact host-pathogen interactions in infectious diseases will open up a new window for developing new tools for disease diagnosis and monitoring. In the current study, the host-pathogen interaction and investigation of the total microbiome and monitoring the changes of microbiome during the infection will be performed in the context of Cryptosporidium and Giardia infected neonatal calves using isolated dung-derived EVs as a non-invasive approach.
EVs are heterogeneous and membrane-enclosed, nano-vesicles naturally released from cells that can mediate various physiological and pathological functions in recipient cells by acting as cargo carriers for molecules such as proteins, lipids, metabolites, and nucleic acids. Since EVs are present in almost all biological fluids, including plasma, urine, seminal fluid, breast milk, serum and faeces, there is a great potential to develop EV-based diagnostic methods that will open up a new window for understanding the host-pathogenic interactions and non-invasive disease investigation. Identification and validation of Cryptosporidium and Giardia virulence factors and pathogenicity are ongoing topics and have not been well defined so far. The main aim of the current study is to understand them based on EV-mediated communication and interaction between host and pathogen.
Proteomics and genomics studies revealed that EVs could be associated with various diseases, abnormalities, cancers, and neurodegenerative diseases in both animals and humans. Therefore, the EVs hold great potential for diagnostics and therapeutics advances in clinical applications. In the past few years, metagenomic analysis of EVs derived from clinical samples has been used to diagnose and monitor the disease, and to identify biological markers in EVs associated with many pathological and physiological conditions. Specifically, finding the interaction of gastric cancer patients and the urine-derived bacterial extracellular vesicles (BEVs) has been conducted by Park et al in 2021 by isolating BEVs presented in urine samples. They were able to identify non-invasive molecular biomarkers for gastric cancer diagnosis. However, in the particular field of metagenomics, where the EVs released by the host, microbiota, and parasite community are present in the digestive tract can be investigated using faeces, and involvement of EVs in Cryptosporidium and Giardia infection diagnosis has not been investigated up to date. To address this challenge, EVs isolated from dung collected from calves with Cryptosporidium and Giardia infections will be analyzed in this study under the wider umbrella of the One Health comparative medicine effort.
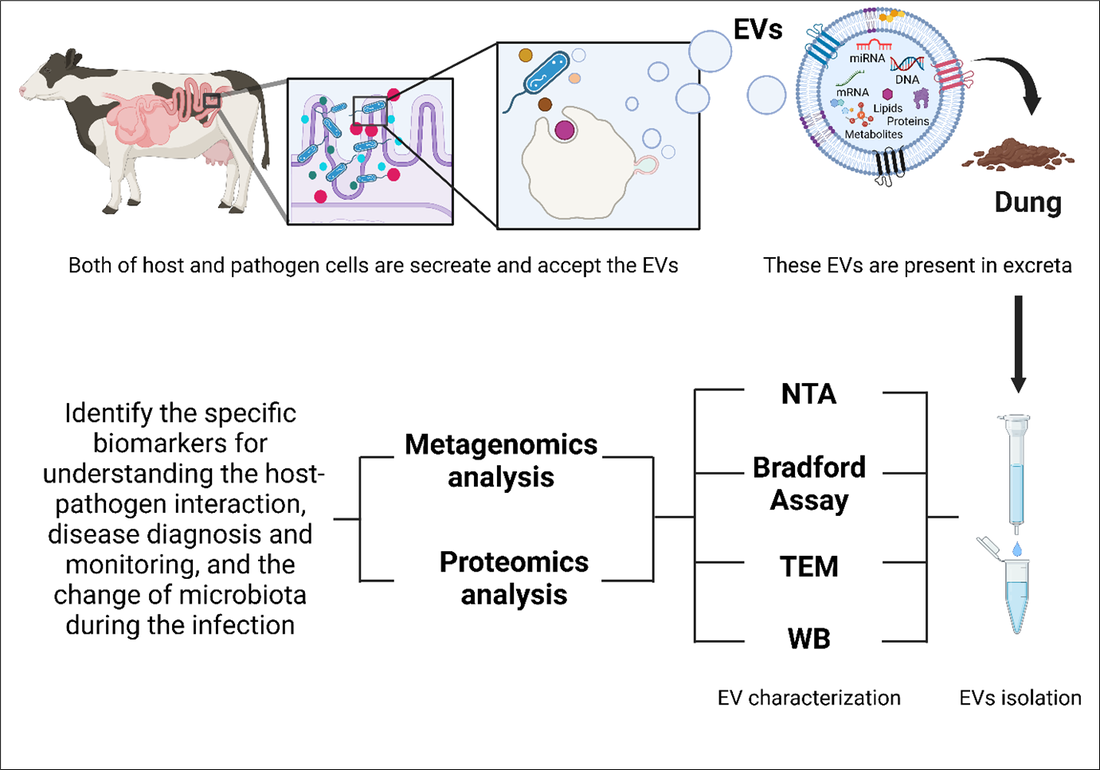
During the course of this project, a methodology will be developed to illustrate the potential of using faeces/dung as a possible non-invasive source for EV isolation and the possibility of using them in understanding the pathophysiology of the disease (Figure 1). In this study, the total genomic profile of faecal-derived EV will be tested. Cattle dung-derived EVs are a combination of nano-vesicles which are originated from different sources including host-derived, parasitic-derived, microbial-derived EVs, and EVs from the diet of the host. Once the genomic profile of EV content is investigated, it will be possible to characterize them and identify their origin. Furthermore, genomic data will be used to discover potential diagnostic molecular biomarkers for understanding the pathophysiology of bovine Cryptosporidium and Giardia infections out of EVs present in dung. A comparative study between the genomics and proteomics data of the EVs isolated from both healthy and infected faecal samples from calves will be performed. This study will aim to understand EVs correlation with disease manifestation in the body, based on the interaction between host and pathogen through host immune response. If successful, this approach may hold important clinical applications and will open a wide variety of opportunities in disease diagnosis and monitoring. Furthermore, this concept can be applied to diagnose and monitor Cryptosporidium and Giardia human infections using human stool samples.
Since people are interacting with animals in their day-to-day life, most zoonotic diseases are commonly found among them. According to the Center for Disease Control and Prevention (CDC, USA), out of 10 known infectious diseases, 6 are transmitted from animals. These types of zoonotic infectious diseases could badly affect not only human and animal health but also the productivity of livestock. Cryptosporidium and Giardia represent the one of the most common intestinal parasites in Eastern Europe and are common in humans and animal species, especially in bovines. Their transmission is typically achieved through the faecal-oral route, by contact with contaminated stool. Cryptosporidium spp. and Giardia spp. have been respectively ranked as the sixth and 11th most important food-borne parasites globally. According to the review by Plutzer et al (2016), upgrading and making the diagnosis/detection procedures are not sufficient to obtain enough information regarding these infectious diseases in Eastern Europe since the variation of diagnostic methodologies, reporting practices throughout the region and not having enough investigation reports.
Conventional diagnostic techniques such as microscopic methods, staining methods, antigen detection tests, immunological/ immunofluorescence assays and nucleic acid based diagnostic methods such as polymerase chain reaction (PCR) are currently being used for the diagnosis of many bovine intestine related/associated infections including cryptosporidiosis (by Cryptosporidium sp.) and giardiasis (by Giardia sp.). Moreover, understanding the exact host-pathogen interactions in infectious diseases will open up a new window for developing new tools for disease diagnosis and monitoring. In the current study, the host-pathogen interaction and investigation of the total microbiome and monitoring the changes of microbiome during the infection will be performed in the context of Cryptosporidium and Giardia infected neonatal calves using isolated dung-derived EVs as a non-invasive approach.
EVs are heterogeneous and membrane-enclosed, nano-vesicles naturally released from cells that can mediate various physiological and pathological functions in recipient cells by acting as cargo carriers for molecules such as proteins, lipids, metabolites, and nucleic acids. Since EVs are present in almost all biological fluids, including plasma, urine, seminal fluid, breast milk, serum and faeces, there is a great potential to develop EV-based diagnostic methods that will open up a new window for understanding the host-pathogenic interactions and non-invasive disease investigation. Identification and validation of Cryptosporidium and Giardia virulence factors and pathogenicity are ongoing topics and have not been well defined so far. The main aim of the current study is to understand them based on EV-mediated communication and interaction between host and pathogen.
Proteomics and genomics studies revealed that EVs could be associated with various diseases, abnormalities, cancers, and neurodegenerative diseases in both animals and humans. Therefore, the EVs hold great potential for diagnostics and therapeutics advances in clinical applications. In the past few years, metagenomic analysis of EVs derived from clinical samples has been used to diagnose and monitor the disease, and to identify biological markers in EVs associated with many pathological and physiological conditions. Specifically, finding the interaction of gastric cancer patients and the urine-derived bacterial extracellular vesicles (BEVs) has been conducted by Park et al in 2021 by isolating BEVs presented in urine samples. They were able to identify non-invasive molecular biomarkers for gastric cancer diagnosis. However, in the particular field of metagenomics, where the EVs released by the host, microbiota, and parasite community are present in the digestive tract can be investigated using faeces, and involvement of EVs in Cryptosporidium and Giardia infection diagnosis has not been investigated up to date. To address this challenge, EVs isolated from dung collected from calves with Cryptosporidium and Giardia infections will be analyzed in this study under the wider umbrella of the One Health comparative medicine effort.
During the course of this project, a methodology will be developed to illustrate the potential of using faeces/dung as a possible non-invasive source for EV isolation and the possibility of using them in understanding the pathophysiology of the disease (Figure 1). In this study, the total genomic profile of faecal-derived EV will be tested. Cattle dung-derived EVs are a combination of nano-vesicles which are originated from different sources including host-derived, parasitic-derived, microbial-derived EVs, and EVs from the diet of the host. Once the genomic profile of EV content is investigated, it will be possible to characterize them and identify their origin. Furthermore, genomic data will be used to discover potential diagnostic molecular biomarkers for understanding the pathophysiology of bovine Cryptosporidium and Giardia infections out of EVs present in dung. A comparative study between the genomics and proteomics data of the EVs isolated from both healthy and infected faecal samples from calves will be performed. This study will aim to understand EVs correlation with disease manifestation in the body, based on the interaction between host and pathogen through host immune response. If successful, this approach may hold important clinical applications and will open a wide variety of opportunities in disease diagnosis and monitoring. Furthermore, this concept can be applied to diagnose and monitor Cryptosporidium and Giardia human infections using human stool samples.
Figure 1. A schematic presentation of the research project I. Nanoparticle Tracking Analysis (NTA), Transmission Electron Microscopy (TEM), Western Blotting (WB).
Project objectives
Using faecal-derived EVs as a non-invasive means for investigating host-pathogen interactions.
Integration with EMU expertise and research strategy and development
The aims and objectives of this project are in line with the aims and objectives of the EMU department of clinical veterinary medicine (CVM) (vl.emu.ee/en/structure/chair-of-clinical-veterinary-medicine/). The CVM research is structured around the areas of antibiotic resistance of pathogenic microorganisms in domestic animals, inflammatory processes in the animals, bovine cattle health, bovine reproductive physiology and diseases, parasitic and orthopedic diseases in small animals, and wild animal anesthesiology. The current research project is well in line with other research projects currently running in the department. One of the current running research projects in the departments is a collaborative research project led by Professor T. Orro, in collaboration with the University of Helsinki to assess the effects of the neonatal immune system development in calves on their health and productivity in cows. The host-pathogen research project integrates very well with this and other projects currently running in the department and will further strengthen the research culture and future prospective of research in the EMU department of clinical veterinary medicine.
Using faecal-derived EVs as a non-invasive means for investigating host-pathogen interactions.
- Optimizing a methodology for isolating EVs from bovine faeces and improving the enrichment efficiency.
- Characterization of EVs present in both healthy and Cryptosporidium, Giardia infected faeces.
- Identifying the molecular biomarkers for diagnosing and monitoring parasitic infections of Cryptosporidium and Giardia using EVs extracted from the faecal samples of calves.
- Elucidating the origin and pathophysiological role and host-pathogens interactions using EV cargo-based biomarkers in relation to infection manifestation.
Integration with EMU expertise and research strategy and development
The aims and objectives of this project are in line with the aims and objectives of the EMU department of clinical veterinary medicine (CVM) (vl.emu.ee/en/structure/chair-of-clinical-veterinary-medicine/). The CVM research is structured around the areas of antibiotic resistance of pathogenic microorganisms in domestic animals, inflammatory processes in the animals, bovine cattle health, bovine reproductive physiology and diseases, parasitic and orthopedic diseases in small animals, and wild animal anesthesiology. The current research project is well in line with other research projects currently running in the department. One of the current running research projects in the departments is a collaborative research project led by Professor T. Orro, in collaboration with the University of Helsinki to assess the effects of the neonatal immune system development in calves on their health and productivity in cows. The host-pathogen research project integrates very well with this and other projects currently running in the department and will further strengthen the research culture and future prospective of research in the EMU department of clinical veterinary medicine.
Project team
|
Egodawatte Gedara Chanaka Nishan Premathilaka
Institute of Veterinary Medicine and Animal Sciences Student |
Alireza Fazeli
Institute of Veterinary Medicine and Animal Sciences Supervisor |
|
Suranga Kodithuwakku
Institute of Veterinary Medicine and Animal Sciences Supervisor |
Toomas Orro
Institute of Veterinary Medicine and Animal Sciences Supervisor |
Septimiu Radu Ionescu
Institute of Veterinary Medicine and Animal Sciences
Supervisor
Institute of Veterinary Medicine and Animal Sciences
Supervisor